We have a significant interest in developing novel proteomic methods that enable a deeper profiling of the cancer "surfaceome", and then developing new therapeutic targets based on these.
Our major published work describes the technology of "structural surfaceomics", enabling discovery of conformation-selective tumor antigens through a combination of cross-linking mass spectrometry and cell surface proteomics (Mandal et al, Nature Cancer (2023)).
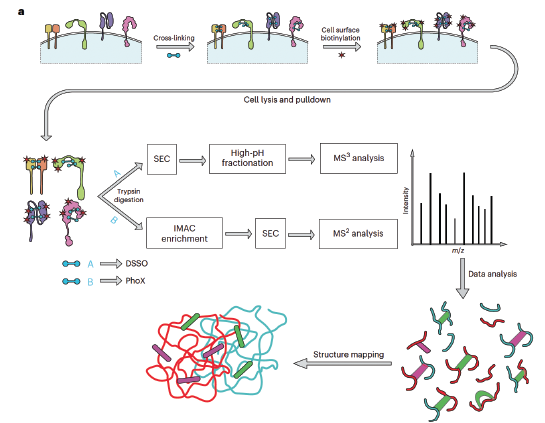
To further enable this technology, we have developed a software, AlphaCross-XL, allowing for rapid mapping and visualization of experimentally-determined crosslinks (Shenoy et al, BioRxiv (2025)).
We are currently exploring other proteomic-based technologies to reveal the "invisible" surfaceome, to identify novel immunotherapeutic targets that cannot be identified with other target discovery strategies.
A major focus of the group is developing protein engineering strategies to recognize these novel surface antigens and then turning these into immunotherapies. We are employing both traditional antibody engineering strategies as well as emerging computational methods for de novo protein design.
